- Human health in the age of genomic medicine
- What is genetic testing
- How are genetic and genomic testing used in human health?
- Clinical genetic testing
- Drug discovery and development
- Consumer genetic testing
- What are the main categories of genetic testing?
- Molecular genetic testing
- Cytogenetic testing
- Biochemical genetic testing
- When are DNA microarrays used in genetic testing?
- Why is NGS revolutionary?
- How is NGS transforming healthcare and clinical practice?
- Oncology
- Transplantation therapy

Genetic testing today and tomorrow
How innovations in genomics and NGS are impacting the lab 
Content
- Microbiome profiling
- Infectious disease and antibiotic resistance
- NGS: bringing precision medicine within reach
- What is NGS library prep?
- Why library prep is key for NGS Success
- Why automate NGS library prep?
- Pain points in NGS library prep
- How automation boosts productivity and quality
- What to consider when choosing an automation solution
- Before you automate
- Automation checklist
- Streamlined solutions from Tecan
- DNA-Seq librararies in 3 simple steps
- A holistic solution for “walk-away” NGS library prep
Human health in the age of genomic medicine
Genetic testing and genomic studies are helping people around the world lead healthier lives and fight disease and infection on a global scale. Exponential advances in high throughput genomic technologies and ‘big data’ analysis have powered large-scale data initiatives such as the 1000 Genomes Project, the 100k Wellness Project, and many others. This explosion of data and technology has enabled development of more rapid and scalable tests that can survey entire genomes and accurately pinpoint genetic changes to a single base pair. As a result, the emerging discipline of genomic medicine, which makes use of personal genomic information to enable more individualized care, is transforming clinical practice.
To keep up with rapidly evolving trends in genomic medicine and human health sciences, an understanding of the basic concepts, technologies and applications used in genetic testing and genomic analysis is essential. Here we address some of the most frequently asked questions on this topic, and take a closer look at some of the fields where next generation sequencing (NGS) is already making an impact on human healthcare and clinical practice.
What is genetic testing?
In its broadest sense, genetic testing refers to direct or indirect analysis of genetic variations and changes in sequence, structure or expression of heritable genes. Depending on the purpose, genetic tests can be carried out on virtually any biological sample that contains intact genetic material, gene products, or related metabolites.
In clinical and regulatory contexts, the term genetic testing is typically reserved for tests that focus on inheritance of a particular trait or single gene—for example, to assess whether an individual has an increased likelihood of inheriting a certain single-gene disorder like cystic fibrosis or Huntington disease. Genetic testing is considered distinct from genomic testing, which involves analysis of changes across a panel of genes or an individual’s entire genome – for example, when assessing a tissue biopsy to determine which genes are driving tumor growth in a cancer patient.
With the advent of higher throughput, larger-scale technologies such as DNA microarrays and massively parallel NGS approaches, where entire gene panels and even whole genomes can be analyzed in a single run, the distinction between genetic and genomic testing is becoming less clear1, and these terms are sometimes used interchangeably.
How are genetic and genomic testing used in human health?
Thousands of human disorders and diseases are now known to result from specific genetic alterations or have a strong genetic component. DNA sequence variations inherited or acquired as a result of diet, lifestyle or environment can determine whether we are susceptible to a particular disease, pathogen or allergen. Our genes also have an impact on how we respond to various medical interventions and treatments, including vaccines, blood transfusions, organ transplantations, pharmaceutical drugs, and cancer therapies—to name just a few.
Genetic testing and genome-wide analyses are therefore informative for diverse applications in human health, and across many disciplines.
Clinical genetic testing
In clinical practice, healthcare practitioners order genetic testing for many purposes, including:
- Pre-natal testing to evaluate risk of genetic disorders or birth defects
- Newborn screening for recessive genetic disorders
- Carrier testing to determine if an individual—usually a prospective parent—carries a specific autosomal recessive disease gene
- Predictive testing to assess predisposition to particular diseases
- Diagnostic testing to diagnose, confirm, classify, stage or rule out a particular genetic condition, before or after symptoms appear
- Treatment monitoring to evaluate efficacy of treatments such as chemotherapy
- Companion diagnostic testing to test whether an individual expresses the target for a particular drug or therapy
- Pharmacogenetic or pharmacogenomic analysis to help in selecting an appropriate dosage or drug
The US Food and Drug Administration (FDA) website for in vitro diagnostics (IVDs) currently lists over 100 nucleic acid-based human genetic tests that have been approved for clinical use2. Approved tests include those for many types of cancer, coagulation factors, neurodegenerative diseases such as Alzheimer’s and Parkinson’s, cystic fibrosis, drug metabolizing enzymes, immune disorders and cardiovascular disease.
Drug discovery and development
Genetic and genomic studies are helping biomedical researchers in drug discovery unravel disease mechanisms, identify novel targets for new drugs and therapies, develop precision medicines, and much more. Genomic analysis, in particular, NGS-based approaches, are informing every stage of the drug discovery process, from basic research and target identification through to clinical trials (Figure 1).
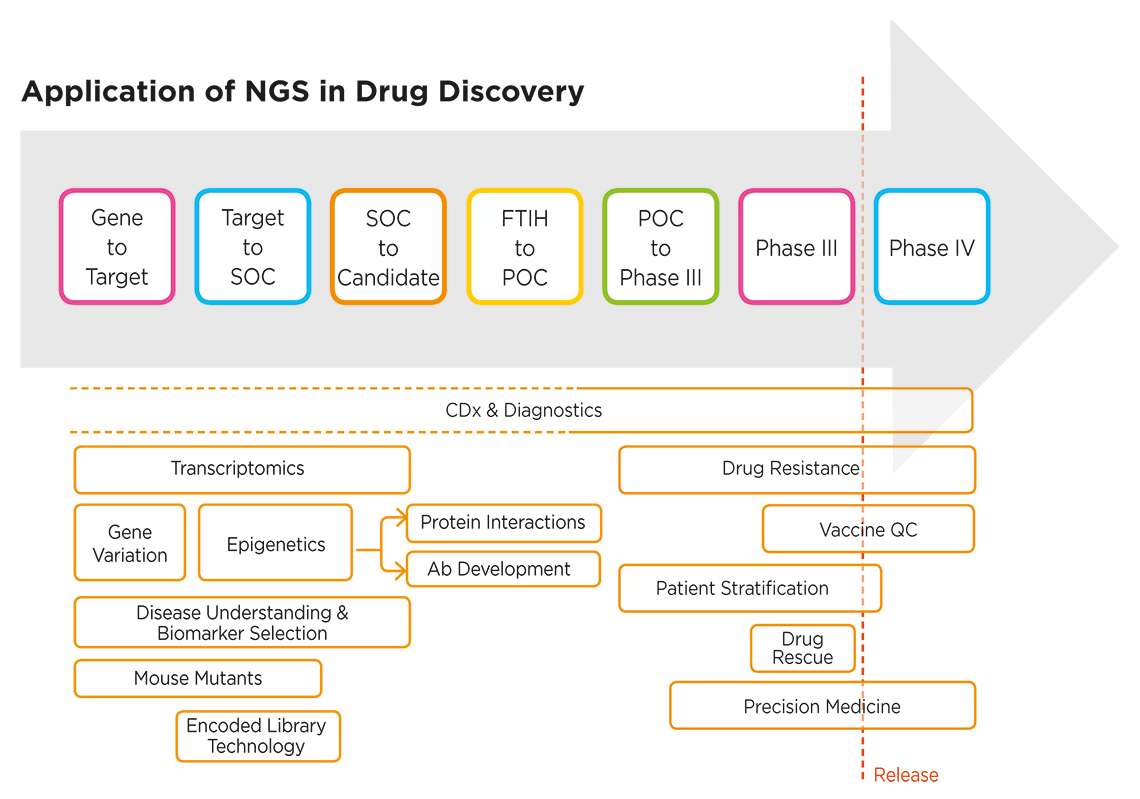
Figure 1. NGS applications extend across the entire drug discovery process3. SOC: standard of care; POC: proof of concept; FTIH: first time in human; CDx: companion diagnostics; QC: quality control.
The use of “omics” analysis approaches—including NGS-based tools for transcriptomics, epigenomics and metagenomics—is enriching our understanding of the complex dynamics and factors underlying human health and disease, which in turn helps fuel the drug discovery process with novel biomarkers. NGS is also an enabling technology for companion diagnostics (CDx), which are increasingly being co-developed from the earliest stages of the drug discovery process.
Consumer genetic testing
Consumers can now purchase affordable DNA tests online, collect their own samples, and receive the results directly. Also known as direct-to-consumer (DTC) genetic tests, home-use DNA tests are providing unprecedented access and insight into our genetic make-up and predisposition to disease. In April 2017, a landmark decision announced by the FDA allowed marketing of the first DTC genetic health risk (GHR) tests for 10 diseases (Table 1). A large number of non-clinical home-use DNA tests are now available from many different providers. While many of them are purchased to gain insight into health, they are also used for purposes as diverse as ancestry research, paternity testing, and tracing absent parents.
| Test target | Description |
| Parkinson’s disease | A nervous system disorder impacting movement |
| Late-onset Alzheimer’s disease | A progressive brain disorder that destroys memory and thinking skills |
| Celiac disease | A disorder resulting in the inability to digest gluten |
|
Alpha-1 antitrypsin deficiency |
A disorder that raises the risk of lung and liver disease |
| Early-onset primary dystonia | movement disorder involving involuntary muscle contractions and other uncontrolled movements |
| Factor XI deficiency | A blood clotting disorder |
| Gaucher's disease (type I) | An organ and tissue disorder |
| Glucose-6-phosphate dehydrogenase deficiency (G6PD) | A red blood cell condition |
| Hereditary hemochromatosis | An iron overload disorder |
| Hereditary thrombophilia | A blood clotting disorder |
Table 1. The first FDA-authorized direct-to-consumer genetic health risk tests, marketed by 23andMe. DNA isolated from a saliva sample is tested for the presence or absence of more than 500,000 genetic variants, some of which are used to assess risk of developing these 10 diseases or conditions4.
Applications for genetic testing continue to grow as new technologies make it easier and more affordable to analyze genetic changes at increasingly high resolution, and across entire genomes and populations.
What are the main categories of genetic testing?
This century’s explosion in genetic testing and genome-wide analysis has been fuelled by continued development of more sensitive, rapid and cost-efficient methods of analyzing genetic material. In particular, chip-based microarray approaches and NGS technologies have enabled more affordable and comprehensive genome-wide analyses at increasingly high resolution. As a result, genetic testing is more accessible and more widely used than ever before.
Genetic testing methods of choice vary according to the type of information required, the level of sensitivity needed, and the time it takes to get results. Clinical genetic testing approaches fall into 3 main categories: molecular, cytogenetic and biochemical:
Molecular genetic testing
Molecular genetic tests detect genetic and epigenetic changes and variations at the molecular level. DNA or RNA is analyzed using techniques such as polymerase chain reaction (PCR), microarrays, Sanger sequencing and NGS. Depending on the methodology, molecular diagnostics can resolve genetic changes at the single nucleotide level (the highest resolution possible). Molecular diagnostic techniques may be used to analyze just a few selected loci or to survey entire exomes or genomes.
Cytogenetic testing
Cytogenetic tests analyze DNA at the chromosomal level to identify gross changes such as additions or deletions of entire chromosomes or long stretches of DNA. Conventional cytogenetic approaches such as karyotyping are giving way to molecular cytogenetic methods, for example chromosomal microarrays (CMAs) or tissue microarrays (TMAs) combined with fluorescence in situ hybridization (FISH) to identify submicroscopic abnormalities not detected by traditional approaches.
Biochemical genetic testing
Biochemical genetic tests measure the downstream effects of genetic changes, rather than changes in the genetic material (DNA or RNA) itself. In this sense, such tests are classed as “indirect”. Biochemical genetic tests typically quantify the activity or levels of gene products (proteins) present in complex biological samples such as urine, blood, cerebrospinal fluid and amniotic fluid. A wide variety of detection techniques and modalities are employed including protein microarrays, HPLC, tandem mass spectrometry (MS/MS), GC/MS, fluorescence and bioluminescence.
When are DNA microarrays used in genetic testing?
With the emergence of DNA microarray chip technology in the mid-1990s, it became possible to analyze thousands of genetic markers in a single run.
Microarray-based gene expression profiling (GEP) identifies disease-associated changes in gene expression by analyzing differential hybridization of cDNA to panels of target sequences arrayed on a chip. Microarray technology can also be applied to microRNA (miRNA) profiling, for example in cancer classification and earlier identification of carcinoma of unknown primary (CUP) origin.
Clinicians and researchers are increasingly making use of microarray-based diagnostics to guide decision-making, especially for challenging samples and complex diseases, where global expression analysis can inform diagnosis, sub-typing, staging and response assessment.
Microarray technology is also being applied for genome-wide analysis of single nucleotide polymorphisms (SNPs). SNP microarray genotyping provides a cost-efficient means of assessing the risk for multiple genetic disorders in a single test. Many providers now offer SNP genotyping as a platform for predictive and pre-symptomatic testing for a wide range of diseases and other conditions. NGS-based SNP genotyping (Genotyping by Sequencing, GBS) is also possible, and in certain applications may offer advantages over microarray approaches, although its use is currently more prevalent in agriculture than human health.
Why is NGS revolutionary?
Completing the first human genome reference sequence took 13 years and around $1 billion. With the latest NGS solutions and advanced bioanalytics, it is now feasible to obtain whole-genome test results in under a day14,15, and the cost of NGS continues to plummet as we enter the era of the $1000 genome16. NGS is without doubt a game changer in genomics and applied fields, including precision medicine and companion diagnostics.
While NGS technologies vary, what they have in common—and what sets them apart from conventional Sanger sequencing as truly disruptive “next generation” solutions—is their ability to run unprecedented numbers of sequencing reactions in parallel on a micro scale that uses far less starting material. The result is significantly faster and more cost-efficient sequencing.
Capable of generating gigabytes of sequence data per day, NGS has fueled an explosion in our genomic knowledge base, adding substantially more breadth and depth to our understanding of health and disease in the process.
Initially NGS DNA sequencing (DNA-Seq) methods were applied to improve the scale and speed of whole genome sequencing (WGS). The power and practical utility of NGS have continued to grow as the technology has been extended to enable longer reads and include more modalities.
NGS technology platforms can now be used for:
- Whole genome sequencing (WGS)
- Targeted panel testing (amplicon- and capture-based approaches)
- SNP calling and targeted genotyping by sequencing (tGBS)
- Whole exome sequencing (WES)
- RNA sequencing (RNA-Seq, miRNA-Seq)
- Chromatin immunoprecipitation (ChIP-Seq)
- Methylation sequencing (Methyl-Seq)
- Regulatory analysis (CLIP-Seq)
- Metagenomics and microbiomics
- Liquid biopsy
- Single-cell sequencing
NGS-based WGS and WES methods have already starting to transform the way various cancers are treated, and have enabled diagnosis of novel genetic disorders. Information from large-scale genome sequencing projects is expanding known genetic profiles across more populations, paving the way for more practical clinical application of genome-wide analyses in disease diagnosis and personalized medicine.
Compared to microarray approaches for gene expression analysis, NGS RNA-Seq offers greater resolution and overcomes array ‘design bias’. NGS can also provide a more complete genomic picture in applications such as SNP genotyping and methylation analysis, rather than being limited by the capacity of the microarray.
In the context of the clinic, NGS offers a significant advantages over conventional gene-by-gene sequencing approaches, particularly when applied to identifying mosaicism in disease diagnosis or elucidating the genetic basis of complex phenotypes, such as those manifested in autoinflammatory diseases, which can be caused by many different genes. NGS methods also offer the potential of higher resolution in cytogenetic testing, although microarrays are currently more cost-efficient and therefore more widely used for such applications.
How is NGS transforming healthcare and clinical practice?
The rapid evolution of sequencing technology in the 21st century has given researchers unprecedented power to understand human health and disease at the biomolecular level, discover better therapeutic targets, and transform clinical care. The following are a few key areas where NGS technology is having significant impact in biomedical research, healthcare and clinical practice.
Oncology
Cancer is one of the leading causes of death worldwide, and has profound impacts on society and healthcare systems at large.
Estimated national expenditures for cancer care in the United States in 2017 were $147.3 billion. In future years, costs are likely to increase as the population ages and cancer prevalence increases5.
A range of biochemical, molecular genetic and cytogenetic methods are used in oncology testing. Newer methodologies, including DNA microarrays and NGS, are improving productivity, accuracy and sensitivity in many oncology applications including susceptibility testing, diagnostics and classification of cancer type.
Targeted re-sequencing for mutation detection and RNA-Seq for gene fusion identification and detection are some of the areas where NGS offers technology advantages in oncology.
Targeted gene panels are often the method of choice in NGS-based oncology testing and risk assessment, largely due to their convenience, cost-efficiency, and high coverage of regions of interest.
The Ovation Fusion Detection BaseSpace Application in conjunction with Tecan’s comprehensive fusion panel provides a simple start to finish workflow. This solution helps us to not only identify clinically actionable fusions but also to perform basic research to study how other fusions may be relevant to cancer progression and prognosis in a single assay.
Cofounder and CEO Veracyte®
Targeted gene panels may comprise in the range of 20 to 500 genes, and fall into two categories:
- Pan-cancer panels enable simultaneous risk assessment of multiple cancer types, such as breast, prostate, colorectal, melanoma, etc. Organ or tumor type-specific cancer panels are useful in cases where the type of cancer susceptibility is known, but more detail is required to reliably assess risk – for example, when there is a family history of the same type of cancer and family members have previously tested negative for common risk factors.
Formalin-fixed, paraffin-embedded tissue (FFPE) is a common source of material for targeted sequencing in cancer studies. Formalin fixation and the small number of relevant cells in many cases can make it challenging to extract sufficient intact nucleic acid for library preparation. High quality DNA with A280:A260 ratios in excess of 1.8 is recommended, in conjunction with a library preparation system specifically designed for low DNA input.
MELANOMA PROGRESSION STUDY USING FFPE SAMPLES
To prepare DNA from FFPE melanocytic neoplasms for targeted sequencing of 293 cancer genes6, Dr. Shain and coworkers chose Tecan’s Ovation® Ultralow Library System. With a yield ranging from 25 to 250 ng of DNA from 150 samples, they obtained an average sequencing coverage of 281×, which enabled identification of somatic mutations despite the presence of significant stromal-cell contamination.
WES and WGS methods have the potential to improve diagnostic success in more complex cancer phenotypes where the disease is genetically heterogeneous and existing targeted panels would not offer sufficient coverage. WGS has the advantage of being free from the bias inherent in targeted approaches, including WES.
The emerging trend toward sequencing matched tumor and normal DNA samples in cancer diagnostics is one of several drivers for increased volumes of NGS and related workflows in oncology labs. To keep pace with this rising demand for higher NGS throughput, rapid adoption of high-throughput solutions for upstream sample handling and library preparation is crucial.
Simplifying and automating laborious and time-consuming pre-analytical steps prior to NGS can reduce time to results from days to hours—at the same time improving quality and cost-efficiency. Celero DNA-Seq kits with NuQuant® provide a streamlined workflow that significantly reduces processing time and bias in the generation of high quality libraries for DNA-Seq. Celero DNA libraries can be prepared, quantified, normalized and pooled in less than four hours on the Fluent Automation Workstation, compared to more than seven hours for library preparation and qPCR quantification with other kits.
Transplantation therapy
Human Leukocyte Antigen (HLA) genes play critical roles in autoimmune disorders, infectious disease responses, and compatibility of tissue and organ transplants. HLA genotyping is thus an essential tool in clinical applications such as organ transplantation, stem cell therapy and diagnosis of HLA-associated diseases. NGS has the potential to revolutionize HLA testing and biobanking by providing allele-level accuracy in a single high-throughput step. For patients in need of vital transplants, this can mean faster tissue matching, earlier treatment and a better chance of survival.
With new therapeutic technologies and better disease detection, the number of transplant patients and potential donors rises year on year, and with it the burden on registries and testing laboratories to coordinate donor-recipient matching more efficiently, coping with an immense volume of sample processing while keeping costs down. For instance, blood and bone marrow transplants are used to treat more than 70 different diseases including the number one childhood cancer - leukemia.
Some 50,000 patients worldwide receive blood stem cell transplants each year, and their fate may well rest on how quickly clinicians are able to find them a well-matched donor through HLA testing.
In many cases finding that perfect match is far from easy – it requires timely and accurate analysis of a highly complex and variable region of the human genome. More widespread adoption of NGS technology for HLA testing promises to ease this burden. The evolution of more affordable NGS platforms, automated HLA sample prep, high-coverage HLA sequencing panels, and improved informatics solutions is making NGS more practical for routine HLA genotyping applications.
Microbiome profiling
There are at least as many microbes colonizing our bodies as there are human cells, with some reports estimating the ratio to be as high as 10 to 17. Microbes inhabit nearly every part of our bodies and play significant roles in both health and disease. Conditions as diverse as obesity, irritable bowel disease, and even depression have been linked to differences in microbiome composition and genetic diversity.
The human gut microbiome is home to more than 1000 bacterial species encoding over 3 million genes carrying the potential to impact human health.8
The rapidly growing discipline of metagenomics relies largely on high throughput sequencing technology for accurate and comprehensive microbiome profiling. NGS overcomes many of the bottlenecks associated with more conventional analysis methods. In particular, NGS enables analysis of microbes that are difficult or impossible to culture. About half of human gut bacteria, for example, are unculturable.
From exploring the genetic diversity of urban microbial communities to understanding the role of microbiota in women’s reproductive health, the applications for NGS metagenomic analysis are diverse and far-reaching in terms of their potential benefit for human health and prevention of disease.
Common NGS-based methods for microbiome analysis include:
- Targeted sequencing – Targeted or amplicon-based sequencing is often the method of choice to profile microbial species in complex samples such as stool, blood or skin. The bacterial 16S rRNA gene is a particularly convenient target because it contains highly conserved sequences that enable design of universal primers for amplification, as well as variable regions that facilitate discrimination of the many different bacterial species that may be present in a single sample. Because sequencing primers are targeted to specific genomic regions, fewer reads per sample are required compared to whole genome sequencing. On the other hand, amplification bias can be problematic. Automation of 16S library preparation for NGS can help minimize the risk of cross-contamination and human error while enabling processing of up to 96 samples in parallel for improved efficiency and turnaround times.
- Shotgun metagenomics – Shotgun methods capture genome-wide DNA sequence information for all of the organisms present in a sample—including viruses and fungi, which cannot be detected by 16S sequencing.
SCHIZOPHRENIA: SHOTGUN METAGENOMICS
Do microbiomes of schizophrenia patients have telltale differences compared to those of control subjects? Castro-Nallar and colleagues found that levels of Ascomycota, a phylum of fungi, were elevated in schizophrenia patients9. Using Ovation® Ultralow DR Multiplex System for shotgun metagenomics, the team demonstrated significant differences in oropharyngeal microbiota at both the phylum and genus levels. - RNA sequencing (RNA-Seq) – RNA-seq adds another dimension to microbiome profiling by providing information about functional gene expression.
MULTIPLE SCHLEROSIS: MICROBIAL EXPRESSION ANALYSIS
Disease-associated differences in expression levels within microbial sample populations can help uncover novel or unexpected pathogenic agents. Perlejewski and colleagues used Tecan’s Ovation® RNA-Seq System V2 to characterize the microbial composition of cerebrospinal fluid (CSF) from patients with inflammatory demyelinating diseases including multiple sclerosis (MS).10 Analysis of over 441,608,474 reads identified varicella zoster virus as a possible agent of MS pathogenesis in one of 12 patients sampled in the study.
Infectious disease and antibiotic resistance
Infectious diseases are responsible for a staggering number of deaths each year and present a significant healthcare burden worldwide. According to the CDC website, in 2015 the number physician visits for infectious and parasitic diseases in the US alone totaled 16.8 million11. At the same time, the increase of drug resistant pathogens threatens to outpace the development of effective antibiotics and antimicrobial agents.
Traditional approaches to the detection of infectious diseases have relied on direct detection of pathogens, making it both time-consuming and challenging to detect of microbes that are resistant to culture or present at very low levels. Molecular genetic approaches to pathogen detection and medical microbiology, particularly NGS-based metagenomic deep sequencing (MDS), can eliminate the need for culturing and identify multiple pathogens in a single run, from a minute amount of starting material. NGS has the potential to decrease time to diagnosis from weeks to days or even hours, while significantly increasing sensitivity and accuracy.
DETECTING CULTURE-RESISTENT PATHOGENS
Metagenomic deep sequencing (MDS) enables rapid detection of pathogens that are difficult or impossible to culture. In a particularly striking case, Wilson and colleagues used MDS to diagnose a rare form of encephalitis caused by a culture resistant amoeba, Balamuthia mandrallis.12 Tecan’s Ovation® RNA-Seq System V2 facilitated rapid and unbiased detection of B. mandrallis transcripts in cerebrospinal fluid samples. The team report that with adequate staffing, their protocol can be completed in under 48 hours.
Among its varied applications in infectious disease, NGS is an important new tool in the fight to reduce antibiotic resistance. It also plays an increasingly crucial role in the timely identification and control of outbreaks such as Ebola and norovirus. In addition to improving diagnostics, molecular genetic methods such as NGS are providing new insights into the mechanisms of host-pathogen interactions.
NGS: bringing precision medicine within reach
NGS-driven genomic studies are unlocking a wealth of new pharmacogenomic insights that are being used to turn the old ‘one size fits all’ approach to medicine on its head. Rather than try to tailor treatments uniquely to individual patients, precision medicine makes intelligent use of genomic information to understand and predict how individuals will respond to particular drugs and therapies based on their genetic makeup. As NGS becomes more universally available, more comprehensive genomic data can be collected using WGS and WES methods, and integrated into electronic health records and genomic databases. At the same time, large-scale genomic data initiatives continue increase the amount of genomic information available for pharmaceutical and clinical diagnostic applications.
Precision medicine relies on companion diagnostic devices and tests to determine whether a particular drug or therapy is appropriate for an individual patient. As the number of targeted and immune therapies increases, regulatory authorities are increasingly encouraging co-development of companion diagnostics during the drug discovery process (Figure 1).
While the majority of regulatory-approved companion diagnostics and complementary diagnostic tests measure only single or a few markers, the tide is turning. In 2017, the US FDA introduced a streamlined path to approval of NGS-based cancer-profiling tools13. Around the same time, it approved clinical use of the first comprehensive NGS-based diagnostic assays. These assays employ panels of hundreds of cancer-related genes comprising actionable cancer targets for which therapies are already approved or in the pipeline. FDA approval of these new ‘mid-size’ NGS diagnostics means that for the first time comprehensive NGS testing is becoming “standard of care” in oncology—a significant conceptual and practical step forward for precision medicine.
Download here our free eBook "Genetic testing basics..."
What is NGS library prep?
Simply put, the goal of NGS library prep is to convert DNA or RNA samples to shorter double-stranded DNA segments, adding adapter sequences that are compatible with the sequencing method of choice.
The library preparation process starts with DNA or RNA, which must first be isolated from the sample. The higher the yield and quality of material obtained from this sample preparation step, the better and more reliable the downstream results will be.
With sufficient quantities of purified DNA or RNA in hand, the core library preparation steps are performed (Figure 1).
Commercial kits for NGS library prep are now widely available. While the core steps are similar, kit protocols may vary significantly depending on vendor-proprietary technology, as well as on the intended applications and sequencing chemistries.
Figure 1. Core stages of NGS library preparation: (1) DNA or RNA is purified from cells, tissues or fluids; (2) RNA samples are converted to double-stranded DNA (dsDNA) and shorter fragments are generated by mechanical, enzymatic or target enrichment methods; (3) fragments are sized and end-ligated with adapter sequences; (4) libraries are quantified, normalized, pooled and validated, ready for sequencing.
For example, fragmentation and adaptor ligation may be combined, amplicon-based or hybrid-capture target enrichment strategies may be used, and PCR amplification may be required at different points in the process.
Kit choice will therefore have significant knock-on consequences for your library prep workflow—affecting variables such as the necessary sample input, number of protocol steps, order of additions, accessory equipment needed, complexity of the workflow, and the time it takes to complete the protocol (Figure 2).
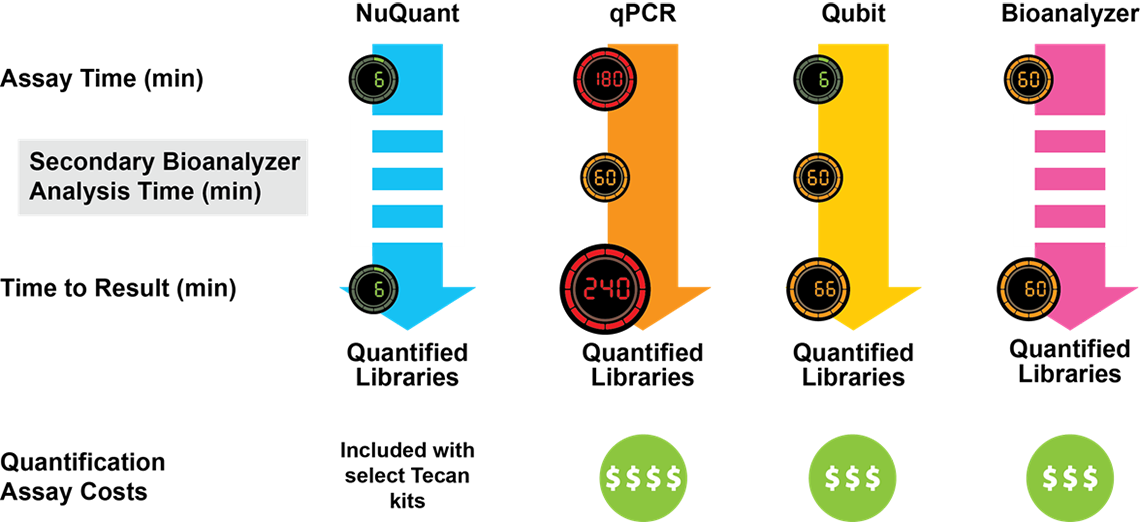
Figure 2. Comparison of NGS Library Preparation protocols. Kits from three different vendors vary with respect to the number of steps, the time (shown in minutes) required for each step, and the total time to completion. Celero DNA-Seq workflow has been streamlined to enable production of sequencing-ready libraries in just 3 hours.
Why library prep is key for successful NGS
No matter which NGS technology or application you are working with, library preparation is the essential foundation for obtaining accurate, reliable and reproducible results from NGS.
Successful NGS is highly dependent on both the amount and quality of the libraries your produce. If you sequence a low-quality library or load an insufficient amount of material onto the flow cell, you risk having an NGS run that does not meet quality control (QC) standards. That means valuable time and resources have been wasted. To make matters worse, in many cases it may not be possible to repeat the work because no more sample is available.
On the other hand, if library irregularities or quality problems go undetected, the consequences can be even more serious. For example, clinical decisions based on sequencing of biased or insufficiently diverse libraries can translate into false-negatives, false-positives and inaccurate diagnoses.
For reliable and reproducible outcomes, NGS libraries must be representative of the sample’s genomic complexity, with even coverage and low bias across the genome.
Why automate NGS library prep?
Introducing inherently high-throughput or “massively parallel” methods such as NGS into the lab often creates unanticipated bottlenecks upstream in the workflow, at the point of DNA/RNA extraction and library prep. This is because manual or semi-automated sample preparation processes are unable to keep pace with the dramatic increase in sequencing capacity, throughput, and necessity of reliability. Not only does automation relieve these bottlenecks, it also reduces error, variability and waste in the process.
Pain points in NGS library prep
Manual library prep is notoriously laborious and time-consuming, with protocols that are highly repetitive and involve hundreds of small-volume pipetting transfers that require high accuracy and precision. Successful results depend on flawless execution at every stage, yet the tedious nature of the work can lead to boredom and lapses in concentration—a recipe for mistakes.
Common “pain points” include:
- Nucleic acid extraction. Conventional methods for DNA and RNA isolation are generally low-throughput and labor-intensive. Solid phase or affinity extraction methods are common, typically using columns or magnetic beads. Repeated equilibration and wash steps can easily go wrong or decrease yield if not performed with care.
- Library construction. Library prep protocols can be complex and demanding, involving a numerous liquid transfers, pipetting of very small volumes, sensitive enzymatic reactions and repetitive clean-up steps. Every single step is critical to success. The entire process can take many hours to complete. Along the way, important variables such as DNA input, molar ratios, incubation temperatures and reaction times must be tightly controlled. The chances of pipetting errors, cross-contamination, process variability and library bias increase when working with large numbers of samples and replicates at small volumes, as is often the case with NGS. Library prep kits are relatively expensive—in some cases costing more than the sequencing run itself—so failures can leave a big dent in your budget.
- Quantification and QC. Accurate library quantification and quality control measures are essential for effective NGS because the sequencing process relies on loading a very precise amount of sample onto the flow cell. If library concentration is too low or too high, the consequences may include run failure or the need for additional sequencing. The most frequently used NGS library QC techniques are qPCR, fluorometry and microfluidic electrophoresis, each of which has one or more drawbacks in terms of accuracy, time or cost.
- Normalization and pooling. When pooling libraries, normalization is crucial to ensure that each library will be equally represented during sequencing. Miscalculations, dilution errors, and pipetting inaccuracies at this stage can jeopardize the sequencing run. Some library prep kits eliminate the need for manual dilution with bead-based normalization. However, if library concentrations are less than around 15nM, bead under-saturation can lead you to over-estimate the concentration, resulting in poor sequencing yields. Manual pooling protocols often instruct the operator to mix libraries thoroughly by gently pipetting up and down up to 10 times. Unless carefully performed, this step can cause you fall at the last hurdle—ending up with degraded sample unfit for sequencing.
How automation boosts productivity and quality
As NGS gets better, faster and cheaper, the biggest productivity and quality gains are now achieved by improving the upstream activities involved in library prep. And the best way to relieve the common bottlenecks and pain points in NGS library prep is to automate the process.
While automation systems were once considered too costly and cumbersome for the average lab, in recent years they have become increasingly versatile, compact and affordable—with reliable solutions that fit most every need and budget.
With the right automation solution in place, you can:
- Deliver more, faster – One of the most obvious benefits of automation is its potential to increase throughput and reduce turnaround times. Multi-channel pipetting and large, open work decks enable you to scale up the number of samples, plates and operations being handled at the same time. Walk-away protocols enable you to maximize operating hours while freeing up staff for higher-level tasks.
- Minimize costly errors and waste – The less hands-on your workflow, the fewer chances there are for people to make mistakes. Compared to humans, robotic systems can reliably run more complex protocols, handle smaller volumes, and process many more samples at a time without accidental mix-ups and pipetting errors. With smaller volumes and fewer experiments that need repeating, you also significantly reduce the amount of reagent and sample waste.
The hidden cost of human error: Operator error can be surprisingly expensive.
Consider, for example, some common error rates in everyday life:
- 1% Wrong character typed
- 2% Wrong vending machine selection
- 6% Wrong entry of 10-digit phone number
If the error rate for something as simple as typing a phone number can be as high as 6%, just imagine how high it might be when carrying out a long, complex, multi-step workflow like NGS library prep.
To find out how much human error could be costing your lab, check out Tecan’s convenient NGS calculator and informative new report: “NGS calculator report: the true cost of sequencing.” - Prevent contamination – In addition to avoiding accidental contamination during manual handling, today’s automation systems have a number of design features that help you proactively prevent contamination during NGS library prep. These include contact-free dispensing, purpose-built protective enclosures, HEPA filtering, UV sterilization, and accessories that are chemically resistant for decontamination and cleaning.
- Improve performance – Today’s automation platforms offer a high degree of accuracy and precision across the many functions required in library prep, including plate handling, dispense head positioning, pipetting, temperature regulation, etc. Liquids can be reliably aspirated and dispensed across the wide range of volumes. Converting from manual to automated library prep also improves performance consistency—from run to run, operator to operator, and lab to lab. This translates into less process variability and higher-quality libraries.
- Ensure traceability and compliance – Sample tracking is one of the benefits of automation that is often overlooked. Automated systems document important run details such as sample ID, protocol name, event timings, reagent lot numbers, volumes, error types, etc. Not only can this information can be invaluable when investigating the root cause of library failures, it also provides clear documentation to demonstrate safety and regulatory compliance, which can be particularly important for labs running clinical tests.
What to consider when choosing an automation solution
While prices have come down, automation systems for library prep are still a big investment. To ensure you get the return you expect, it pays to do your research.
What are the biggest problem areas and obstacles for your lab? Does your budget take into account all the “hidden” indirect costs of NGS library prep? Which features and functionalities are absolute “must-haves”—now and for the future?
To get you started, here are some important considerations and tips from our genomics experts.
Before you automate
- Audit your workflows - Just as important as whether to automate is the question of what to automate. Beware that automation won’t necessarily improve a bad workflow. You may need to optimize and make some protocol changes first. Before implementing any automation solution, we recommend auditing your entire workflow—from sample collection right through to library validation (Figure 3). This will help you pinpoint sources error, waste and inefficiency.
- Think outside the box - While it is tempting to stick with ‘tried and true’ protocols, with a little out-of-the-box thinking you may find you can combine, shorten or even eliminate processing steps. For example: Could use of master-mixes reduce the number of dilution steps and solutions to be prepared? Could alternative technologies or kits eliminate the need for multiple clean-up and wash steps? Could switching to bead-based enrichment methods enable more tasks to be performed on the same workstation?
- Look for opportunities to integrate - Much waste, inefficiency and risk can be eliminated from NGS by integrating as much of the pre-analytical processing as possible. On the other hand, quality and compliance considerations may mean that some workflows need to be kept separate. Knowing which processes you can and cannot integrate will give you an idea of whether you need to budget for more than one automation system, and what types of accessories and configurations will be required for each.
- Consider “hidden” costs – When comparing automation solutions, don’t be tempted to focus on the capital investment alone and lose sight of some of the other factors that affect the overall cost of library prep. A thorough workflow audit will highlight some of the less obvious expenses associated with manual processing or low-budget automation solutions. These may include higher error rates, unnecessary waste, extra time and labor, greater training burden, system downtime, and unplanned service visits. When factors like these are taken into account, you may find that a higher end automation platform will prove more cost-efficient in the long run than semi-automated or low-budget options.
Automation checklist
After auditing your processes and identifying sources of error and inefficiency, you will have a clearer picture of what to look for in a library prep automation solution. When evaluating the different platforms and vendors, here are some important factors to include on your checklist:
- Technical utility and performance: Does the platform have the core liquid handling and robotics capabilities required for your applications? Is it compatible with preferred kits, consumables and reagents? Can you work with a variety of sample holders (e.g. multi-well plates, slides, tubes)? Does the system have the deck capacity and performance specifications you need to meet goals for throughput and turnaround time? etc.
- Integration capability: How easily can you integrate the various procedural steps of your workflow? Will the platform interface with all the necessary supporting equipment such as shakers, magnetic separators, incubators, PCR machines, barcode readers, etc.?
- Flexibility: Will the system be adaptable and scale to meet your changing needs? Will you have the freedom to work with new types of kits, methodologies and chemistries? Can you reconfigure the work deck easily? Can you edit existing protocols and create or import new ones? Is the platform an “open” system that will allow you to run other applications besides library prep, or is it dedicated to a narrow set of procedures and applications?
- Quality and compliance: Are the system specifications appropriate to meet your quality goals? Has the system been designed and demonstrated to comply with the relevant safety and regulatory guidelines for your industry?
- User friendliness: How easy is it to use the system? Is the software interface intuitive? Can you create and edit protocols without needing to know any programming languages? How much training is necessary to operate the system? Will it be easy to train new users? Does the system provide any pre-run checks and visuals to confirm that the run protocol and worktable are set up correctly?
- Service support: What level of service is provided? Is there a 24/7 support line? Are support staff knowledgeable and experienced with genomics applications?
Streamlined solutions from Tecan
In conjunction with smart automation solutions, choosing the right NGS library preparation kit can help you to streamline your process and generate high-quality libraries. Ideally, you should select kits that require the fewest steps, as this decreases the length of your library preparation process while minimizing sample loss and the likelihood of errors. One way to achieve this is to use kits that reduce the number of bead clean-up steps.
DNA-Seq librararies in 3 simple steps
The Celero DNA-Seq library preparation kit is an innovative system that streamlines library preparation for Illumina sequencers. Celero’s addition-only workflow simplifies library preparation with 3 master mixes and only 1 bead purification clean-up following library construction (Figure 4). Elimination of post-ligation bead purification saves time compared to other methods.
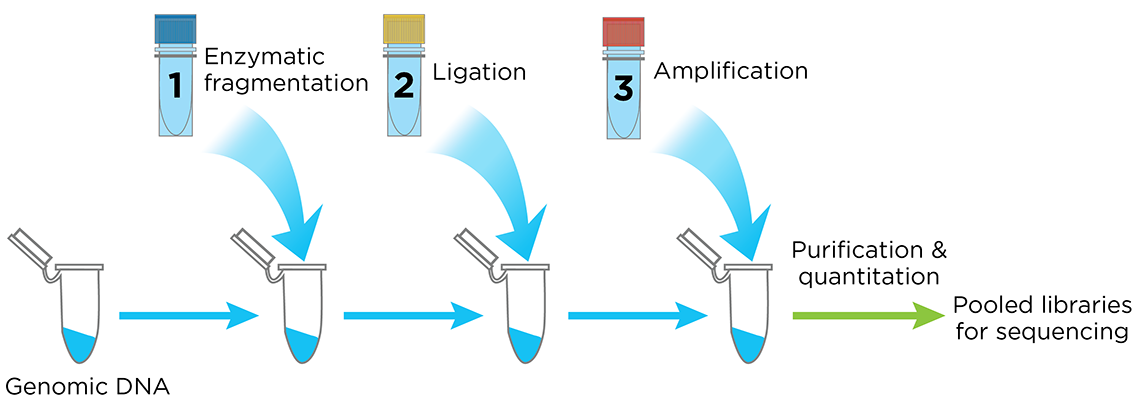
Figure 4. The Celero PCR Workflow. Enzymatic Fragmentation DNA-Seq Kit offers a simple, addition-only workflow for generating quantified libraries ready for sequencing. A separate kit is available for mechanical fragmentation.
Celero DNA-Seq library preparation kit also includes Tecan's breakthrough library quantitation method, NuQuant®, which saves both time and cost in measuring library concentration for pooling. With the proven DimerFree™ technology you can use from 10 ng to 1 μg of intact DNA and greater than 200 ng of FFPE DNA with a single kit without the worry of GC bias, eliminating the need of doing an adapter titration.
Typical DNA-Seq library preparation protocols may have up to 9 separate steps and take over 6 hours to complete. With Celero, you can have sequencing-ready libraries in under 4 hours (Figure 2).
A holistic solution for “walk-away” NGS library prep
Many automated solutions for library prep still require significant hands-on time. This is because they can involve a lot of user input in reagent preparation and loading, and they can’t fully automate the labor-intensive steps involved in QC.
To solve this problem, a fully automated solution with integrated QC has recently been introduced. The system provides an innovative new approach that could help you improve NGS library quality while transforming your workflow efficiencies.
NGS DreamPrep combines the reliable and flexible Fluent Workstation with an integrated Infinite® plate reader, enabling users to transform up to 96 DNA samples into normalized, ready-to-sequence libraries. Designed for compatibility with commercial sequencing kits such as Celero DNA-Seq and Universal Plus mRNA-Seq, NGS Dream Prep brings library preparation, quantification, normalization and pooling of NGS libraries into a single automated workflow that you can control through one easy-to-use software interface (Figure 5).
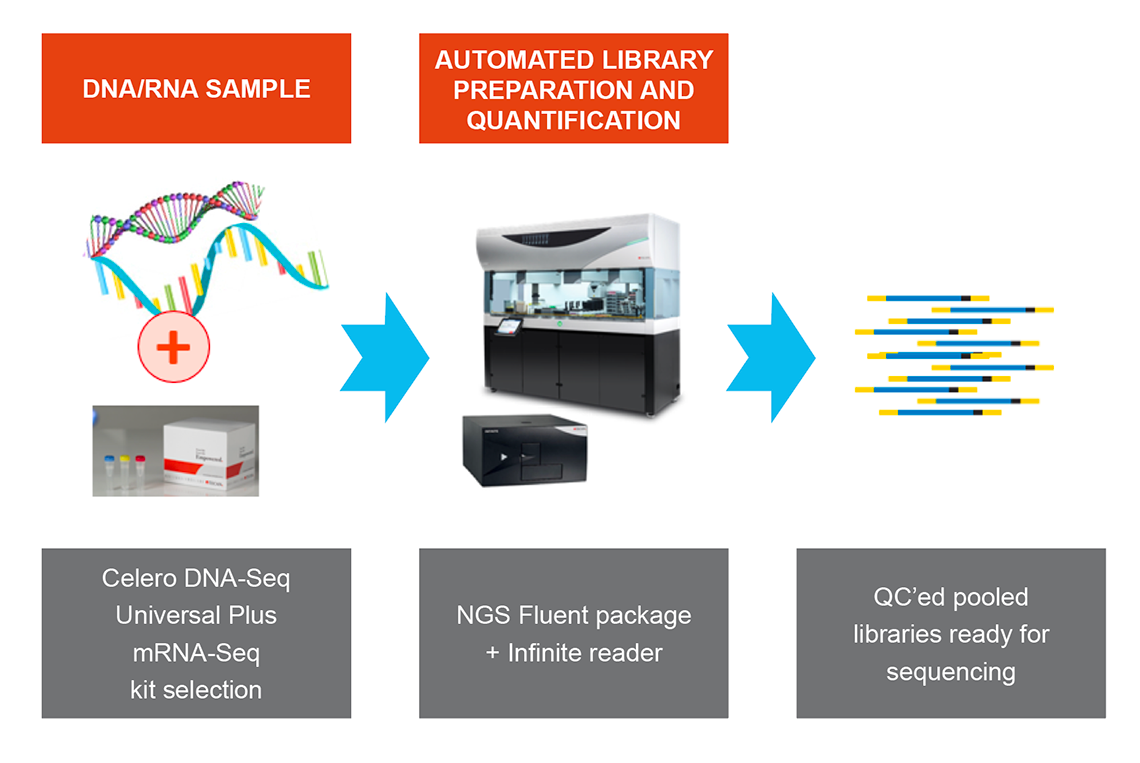
Figure 5. NGS DreamPrep: A holistic solution that integrates library preparation, quantification, normalization and pooling of NGS libraries into a single automated workflow.
Download here our free eBook "Automating library prep..."
1 Katsanis SH and Katsanis N. Molecular genetic testing and the future of clinical genomics. Nature Rev Genet (2015) 14(6):415-426.
2 US Food and Drug Administration Website: Nucleic acid based tests. Page last updated: 11 Sep 2018. Accessed 12 Nov 2018.
3 Adapted from source: Yadav NK, et al. Next Generation Sequencing: Potential and Application in Drug Discovery. Published online in The Scientific World Journal (2014), Article ID 802437, http://dx.doi.org/10.1155/2014/802437.
4 US Food and Drug Administration website: FDA news release 06 Apr 2018. FDA allows marketing of first direct-to-consumer tests that provide genetic risk information for certain conditions. Accessed 12 Nov 2018.
5 National Cancer Institute website: Cancer statistics. Last updated: 27 Apr 2018. Accessed 12 Nov 2018. Accessed 12 Nov 2018.
6 Shain, HA et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med (2015) 373: 1926-36.
7 National Institutes of Health. (2012, June 13). NIH Human Microbiome Project defines normal bacterial makeup of the body. Retrieved from https://www.nih.gov/news-events/news-releases/nih-human-microbiome-project-defines-normal-bacterial-makeup-body
8 Qin, J et al. A human gut microbial gene catalog established by metagenomic sequencing. Nature (2010) 464:59-65.
9 Castro-Nallar, E. et al. Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls. PeerJ (2015) 3: e1140. Published online. DOI: 10.7717/peerj.1140.
10 Perlejewski, K et al. Metagenomic analysis of cerebrospinal fluid from patients with multiple sclerosis. Adv Exp Med Biol (2016) 935:89-98.
11 Centers for disease control and prevention website: Infectious disease. Last updated: 3 May 2017
12 Wilson, MD et al. Diagnosing Balamuthia mandrillaris encephalitis with metagenomic deep sequencing. Ann Neurol (2015) 78:722-730.
13 “FDA unveils a streamlined path for the authorization of tumor profiling tests alongside its latest product action.” U.S. Food & Drug Administration, U.S. Department of Health and Human Services.
14 Collins, F. (2019, May 7) Whole-genome sequencing plus AI yields same-day genetic diagnoses. NIH Director’s Blog. Retrieved June 20, 2019 from: https://directorsblog.nih.gov/2019/05/07/whole-genome-sequencing-and-ai-yields-same-day-genetic-diagnoses
15 Clark, MM et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci Transl Med (2019) 11 (489). DOI: 10.1126/scitranslmed.aat6177.
16 Rincon, P. (2014, Jan 15) Science enters $1,000 genome era. BBC News website. Retrieved June 20, 2019 from: https://www.bbc.co.uk/news/science-environment-25751958
For research use only.